Abstract
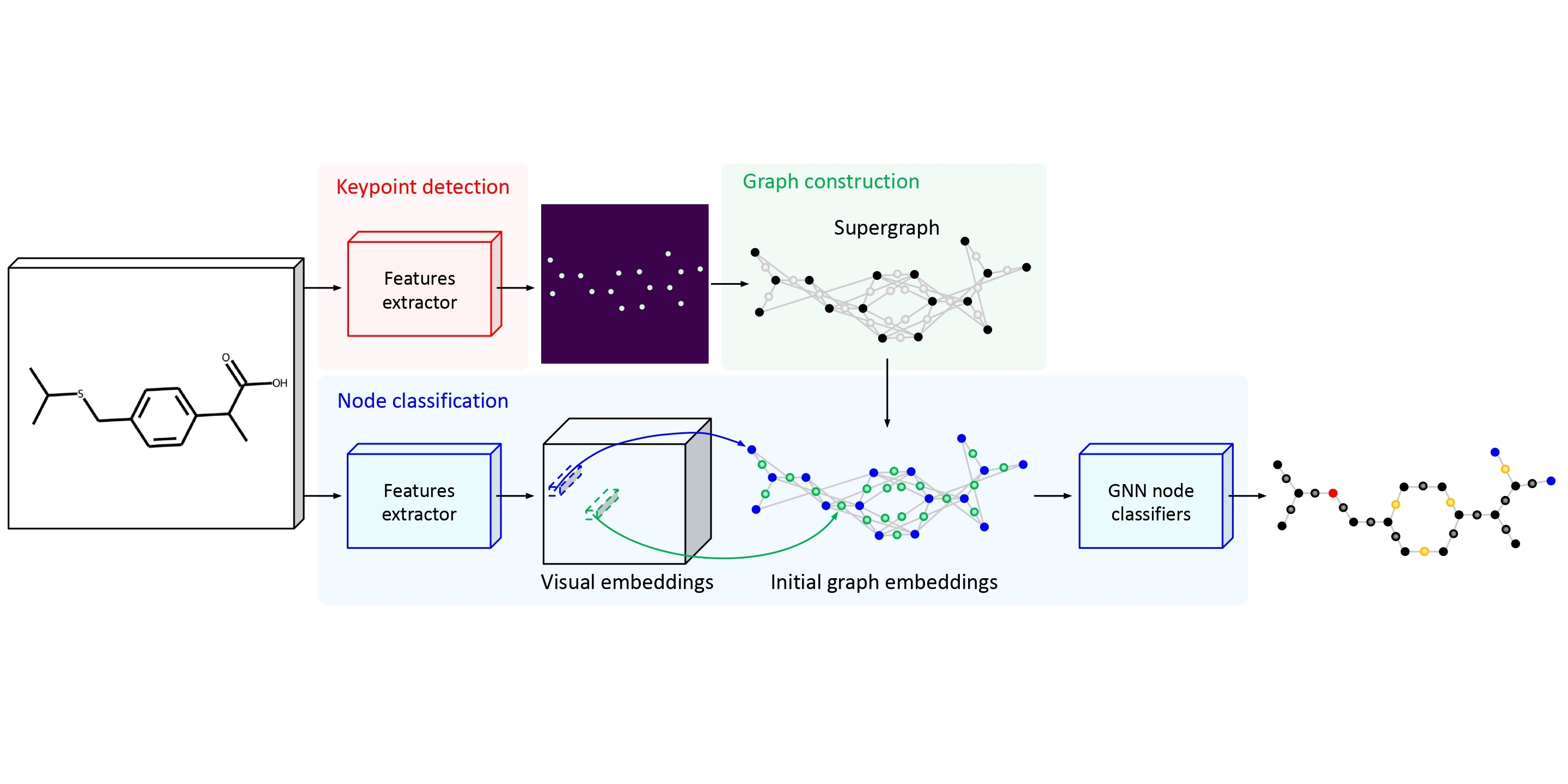
The automatic analysis of chemical literature has immense potential to accelerate the discovery of new materials and drugs. Much of the critical information in patent documents and scientific articles is contained in figures, depicting the molecule structures. However, automatically parsing the exact chemical structure is a formidable challenge, due to the amount of detailed information, the diversity of drawing styles, and the need for training data. In this work, we introduce MolGrapher to recognize chemical structures visually. First, a deep keypoint detector detects the atoms. Second, we treat all candidate atoms and bonds as nodes and put them in a graph. This construct allows a natural graph representation of the molecule. Last, we classify atom and bond nodes in the graph with a Graph Neural Network. To address the lack of real training data, we propose a synthetic data generation pipeline producing diverse and realistic results. In addition, we introduce a large-scale benchmark of annotated real molecule images, USPTO-30K, to spur research on this critical topic. Extensive experiments on five datasets show that our approach significantly outperforms classical and learning-based methods in most settings.
Paper
 | Lucas Morin, Martin Danelljan, Maria Isabel Agea, Ahmed Nassar, Valéry Weber, Ingmar Meijer, Peter Staar, Fisher Yu MolGrapher: Graph-based Visual Recognition of Chemical Structures ICCV 2023 |
Code

github.com/DS4SD/MolGrapher
Citation
@inproceedings{morin2023molgrapher,
title={MolGrapher: Graph-based Visual Recognition of Chemical Structures},
author={Lucas Morin and Martin Danelljan and Maria Isabel Agea and Ahmed Nassar and Valery Weber and Ingmar Meijer and Peter Staar and Fisher Yu},
journal={International Conference on Computer Vision (ICCV)},
year={2023}
}